Reproducible Analysis With R
Oct 16-17, 2018
1 Introduction
Reproducibility is the hallmark of science, which is based on empirical observations coupled with explanatory models. While reproducibility encompasses the full science lifecycle, and includes issues such as methodological consistency and treatment of bias, in this course we will focus on computational reproducibility: the ability to document data, analyses, and models sufficiently for other researchers to be able to understand and ideally re-execute the computations that led to scientific results and conclusions.
1.1 The Reproducibility Crisis
J. P. A. Ioannidis (2005) highlighted a growing crisis in reproducibility of science when he wrote that “Most Research Findings Are False for Most Research Designs and for Most Fields”. Ioannidis outlined ways in which the research process has lead to inflated effect sizes and hypothesis tests that codify existing biases. Subsequent research has confirmed that reproducibility is low across many fields, including genetics (J. P. A. Ioannidis et al. 2009), ecology (Fraser et al. 2018), and psychology (Open Science Collaboration 2015), among others. For example, effect size has been shown to significantly decrease in repeated experiments in psychology (1.1).
![Effect size decreases in replicated experiments [@open_science_collaboration_estimating_2015].](images/effect-size.png)
Figure 1.1: Effect size decreases in replicated experiments (Open Science Collaboration 2015).
1.2 What is needed for computational reproducibility?
The first step towards addressing these issues is to be able to evaluate the data, analyses, and models on which conclusions are drawn. Under current practice, this can be difficult because data are typically unavailable, the method sections of papers do not detail the computational approaches used, and analyses and models are often conducted in graphical programs, or, when scripted analyses are employed, the code is not available.
And yet, this is easily remedied. Researchers can achieve computational reproducibility through open science approaches, including straightforward steps for archiving data and code openly along with the scientific workflows describing the provenance of scientific results (e.g., Hampton et al. (2015), Munafò et al. (2017)).
1.3 Conceptualizing workflows
Scientific workflows encapsulate all of the steps from data acquisition, cleaning, transformation, integration, analysis, and visualization.
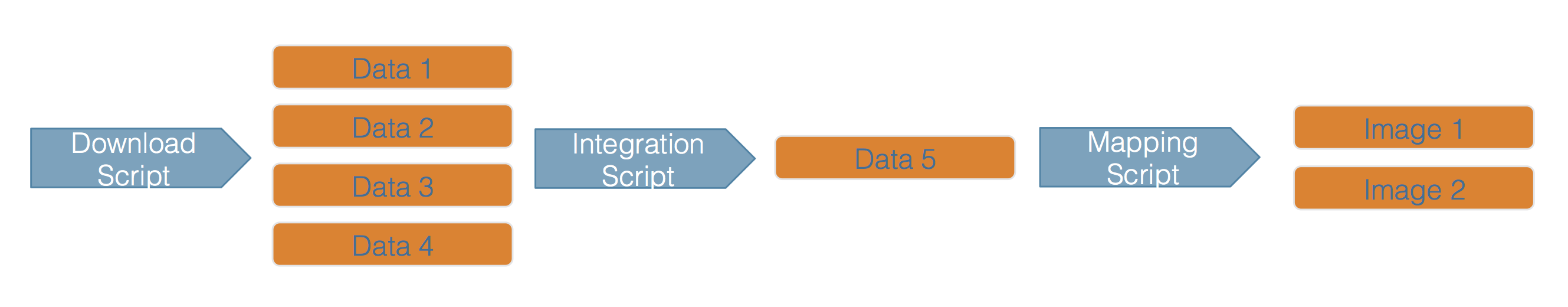
Figure 1.2: Scientific workflows and provenance capture the multiple steps needed to reproduce a scientific result from raw data.
Workflows can range in detail from simple flowcharts (1.2) to fully executable scripts. R scripts and python scripts are a textual form of a workflow, and when researchers publish specific versions of the scripts and data used in an analysis, it becomes far easier to repeat their computations and understand the provenance of their conclusions.
Within many science disciplines, researchers are seeing the power of building reproducible workflows. For example, within fisheries science, the ICES Transparent Assessment Framework (TAF) is being developed to help researchers share stock assessments consistently and completely (1.3).
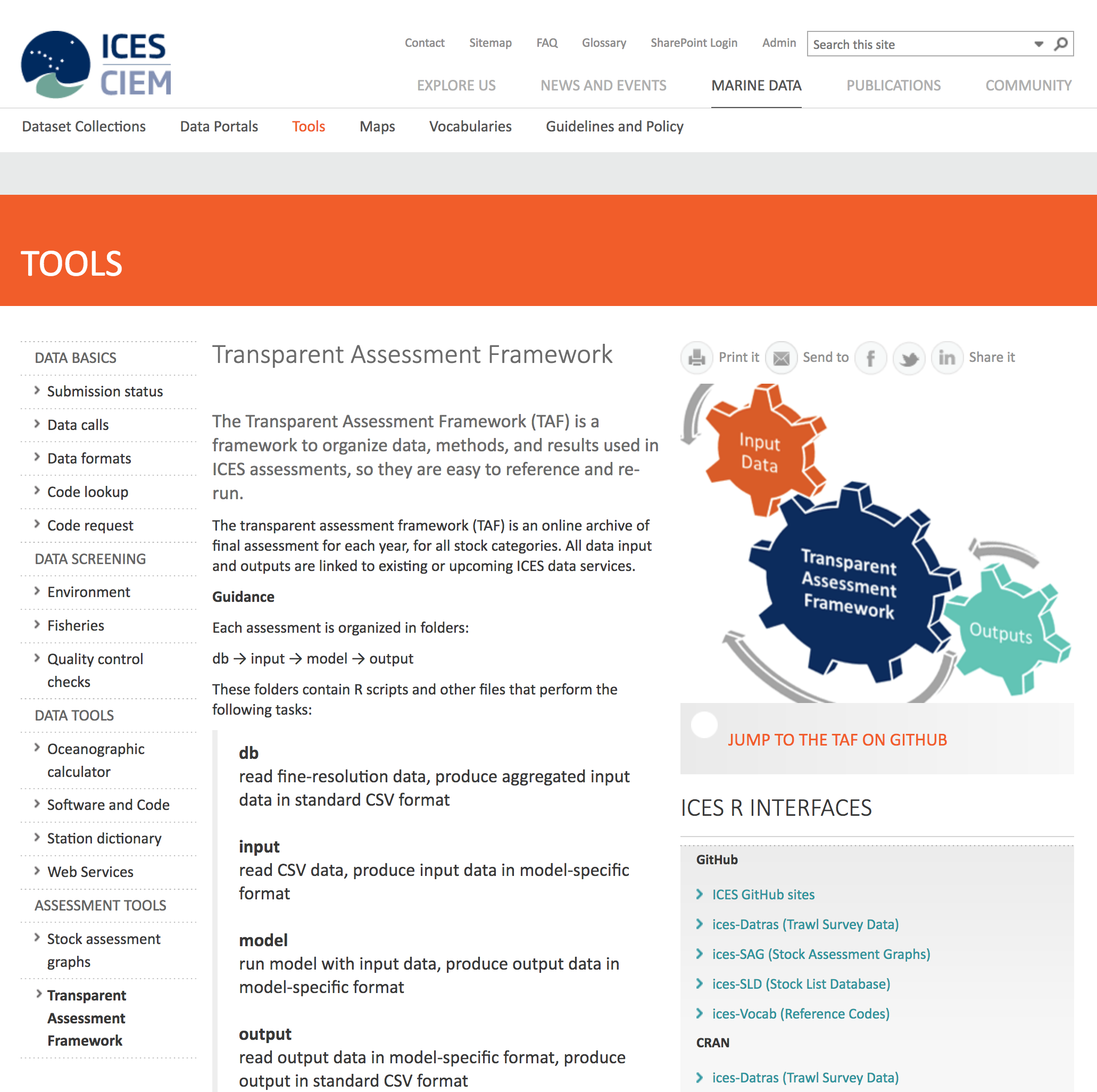
Figure 1.3: ICES Transparent Assessment Framework
1.4 Credit where credit is due
A major advantage of preserving the details of scientific workflows (data, inputs, outputs, and code) is that data citations become so much more effective. Today, driver data sets like global environmental variables, code libraries, and simulated data are critical to many scientific outcomes, but are rarely if ever cited. By preserving scientific workflows, a citation to a highly derived product can be traced back through its chain of provenance to give credit to the researchers and analysts that created those critical precursor products (1.4). Which also helps with a full understanding of just how conclusions were made in an analysis.
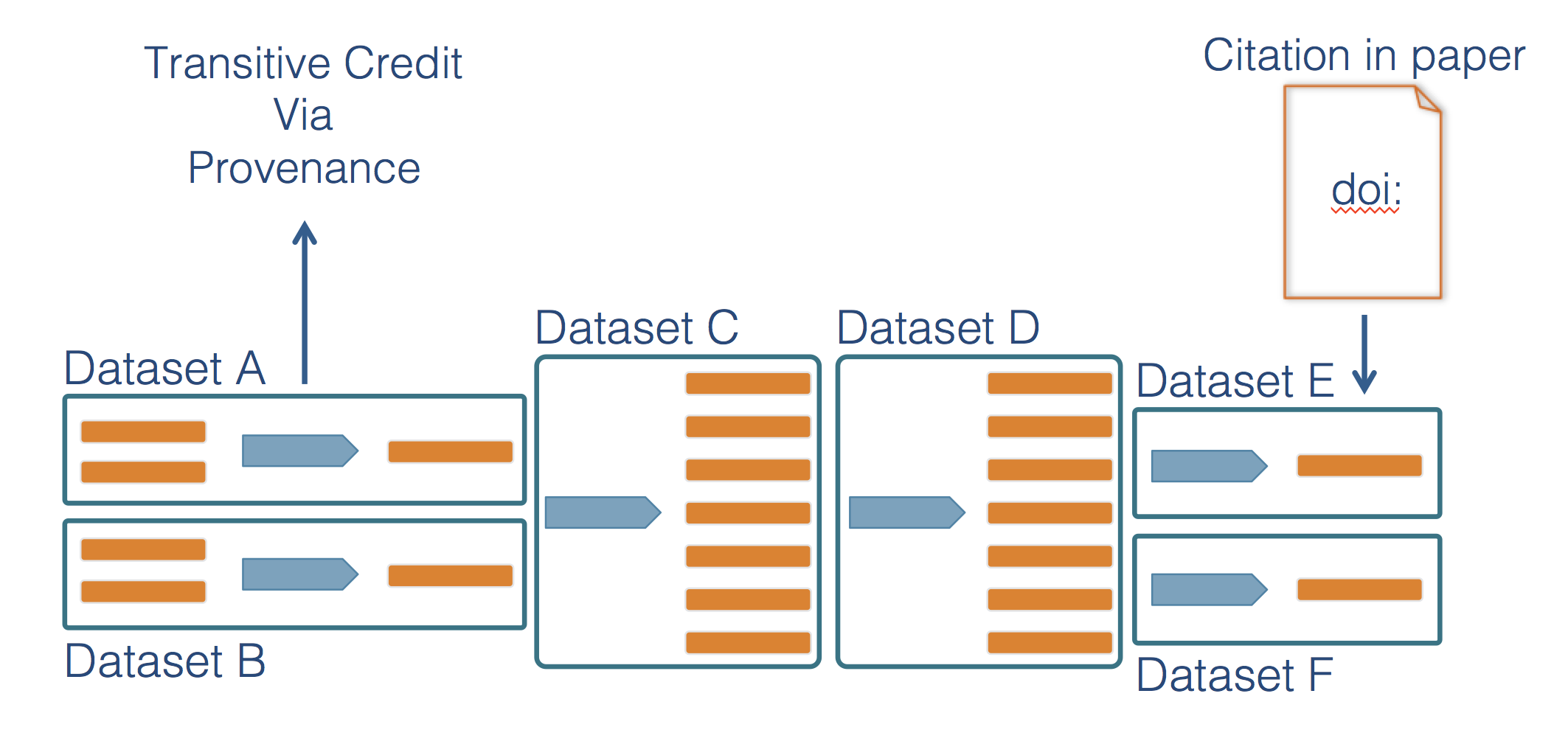
Figure 1.4: Scientific workflows provenance allows credit to be assigned to precursor data and analytical work.
1.5 Course Objectives
This event will cover techniques for building reproducible analysis workflows using the R programming language through a series of hands-on coding sessions. We will use examples from integrating salmon brood data across the state of Alaska to show how heterogeneous data can be cleaned, integrated, and documented through workflows written in RMarkdown. After an overview of the use of RMarkdown for literate analysis, we will dive into critical topics in data science, including version control, data modeling, cleaning, and integration, and then data visualization both for publications and the web. Major topics will include:
- Literate analysis using RMarkdown
- Version control for managing scientific code
- Data modeling, cleaning, and integration
- Publishing data and code
- Data visualization for the web
1.6 Course details
This mini-course will be a hands-on experience, with most of the material presented through a series of tutorials. It is meant as a survey of techniques that will motivate participants to continue self-study in reproducible science.
While prior knowledge of R is a pre-requisite for the mini-course, participants enter with a range of experience with scientific computing in R. We will try to accommodate this diversity, while maintaining a reasonable pace through the materials.
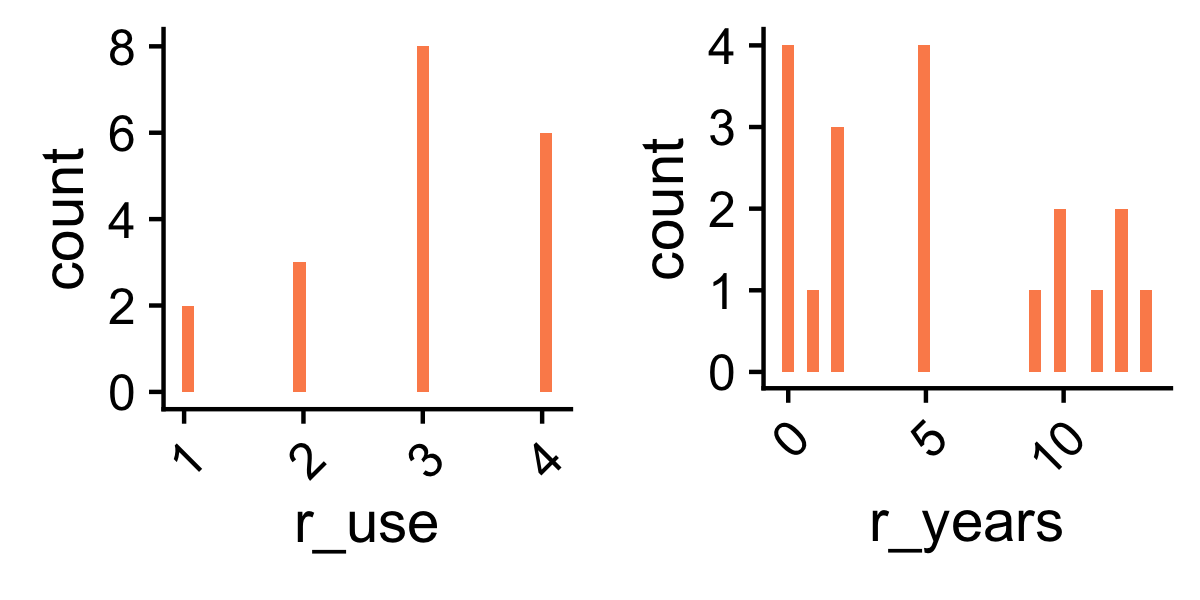
Fig 1.5: Class participants: number of years using R (right) and expertise self evaluation (left).
We will conduct the course in a supportive and friendly manner, working to build skills, confidence, and curiosity about data science. We encourage an informal atmosphere, where questions and discussion are welcome.
References
Ioannidis, John P A. 2005. “Why Most Published Research Findings Are False.” PLoS Medicine 2 (8): e124. doi:10.1371/journal.pmed.0020124.
Ioannidis, John P A, David B Allison, Catherine A Ball, Issa Coulibaly, Xiangqin Cui, Aedín C Culhane, Mario Falchi, et al. 2009. “Repeatability of Published Microarray Gene Expression Analyses.” Nature Genetics 41 (2): 149–55. http://www.ncbi.nlm.nih.gov/pubmed/19174838.
Fraser, Hannah, Timothy Parker, Shinichi Nakagawa, Ashley Barnett, and Fiona Fidler. 2018. “Questionable Research Practices in Ecology and Evolution.” Open Science Framework. doi:10.17605/OSF.IO/AJYQG.
Open Science Collaboration. 2015. “Estimating the Reproducibility of Psychological Science.” Science 349 (6251): aac4716–aac4716. doi:10.1126/science.aac4716.
Hampton, Stephanie E, Sean Anderson, Sarah C Bagby, Corinna Gries, Xueying Han, Edmund Hart, Matthew B Jones, et al. 2015. “The Tao of Open Science for Ecology.” Ecosphere 6 (July). doi:http://dx.doi.org/10.1890/ES14-00402.1.
Munafò, Marcus R., Brian A. Nosek, Dorothy V. M. Bishop, Katherine S. Button, Christopher D. Chambers, Nathalie Percie du Sert, Uri Simonsohn, Eric-Jan Wagenmakers, Jennifer J. Ware, and John P. A. Ioannidis. 2017. “A Manifesto for Reproducible Science.” Nature Human Behaviour 1 (1): 0021. doi:10.1038/s41562-016-0021.