Downloading packages from the Arctic Data Center
Dominic Mullen
Last updated 2018-12-12
d_download_ADC_packages.RmdMac Users:
First install the following R packages:
install.packages("devtools")
devtools::install_github("ropensci/EML")
devtools::install_github("NCEAS/datamgmt")
library(datamgmt)
library(dataone)PC Users:
We currently have issues installing datamgmt on PCs. Instead of installing the package, we can load the functions directly into your global environment by copy-pasting. First install the following R packages:
install.packages("devtools")
devtools::install_github("ropensci/EML")
devtools::install_github("NCEAS/arcticdatautils")
install.packages("dataone")
install.packages("magrittr")
library(dataone)
library(magrittr)Source (copy-paste into your R console) the file https://github.com/dmullen17/work-samples/blob/master/R/wg_helpers.R
Note: This concludes the differences between Mac and PC installs.
Next, we need to specify which DataONE Member Node we want to interact with. The following code specifies the Arctic Data Center. Most of the functions that we use specify the mn as the first argument. First, we specify the Coordinating Node and then its corresponding Member Node.
Download a small Data Package
- Click the “Download” or “Download all” buttons on the ADC User Interface.
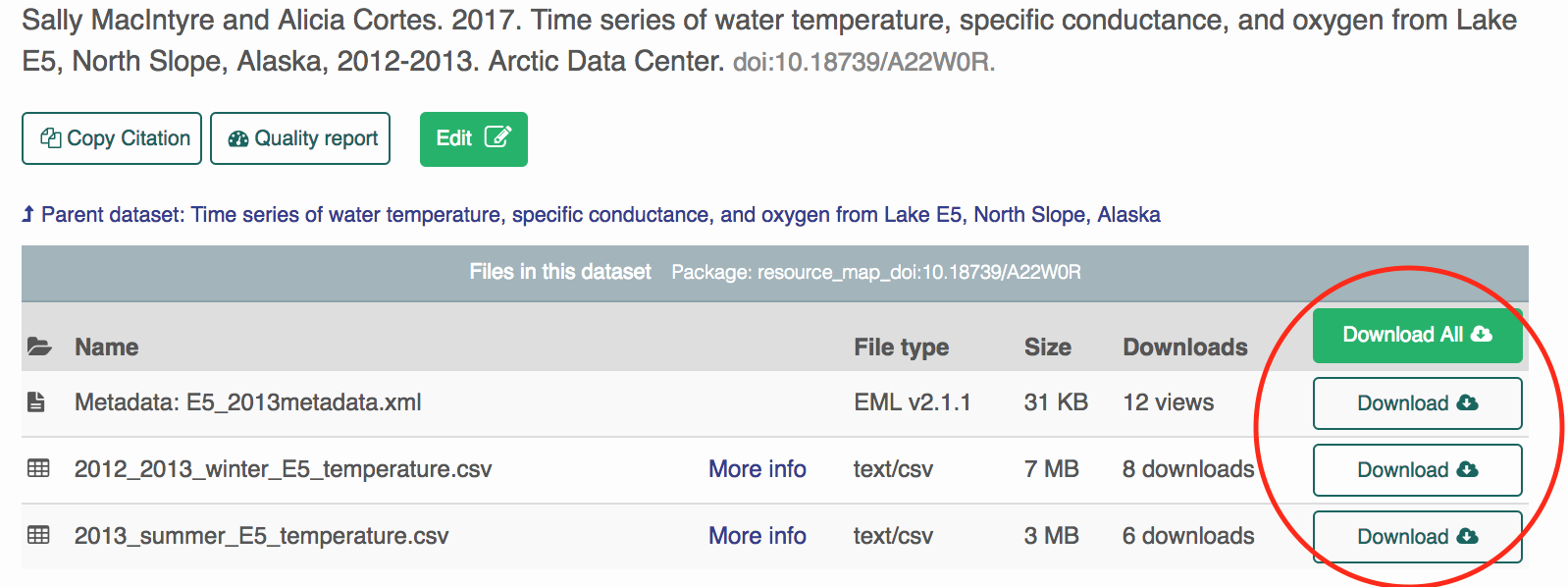
- Download the attribute (column metadata) by pasting the URL into this function. This reads the attributes into a list of data.frames by default.
Optional arguments:
-
write_to_csv- writes each data.frame to a csv in the specifieddownload_directory -
prefix_file_names- prepends the DOI identifier to each file -
download_directory- required ifwrite_to_csv = TRUE
attributes <- get_eml_attributes_url(mn,
"https://arcticdata.io/catalog/#view/doi:10.18739/A22W0R",
write_to_csv = TRUE,
prefix_file_names = TRUE,
download_directory = "/home/dmullen/downloads") Download a Data Package without the UI
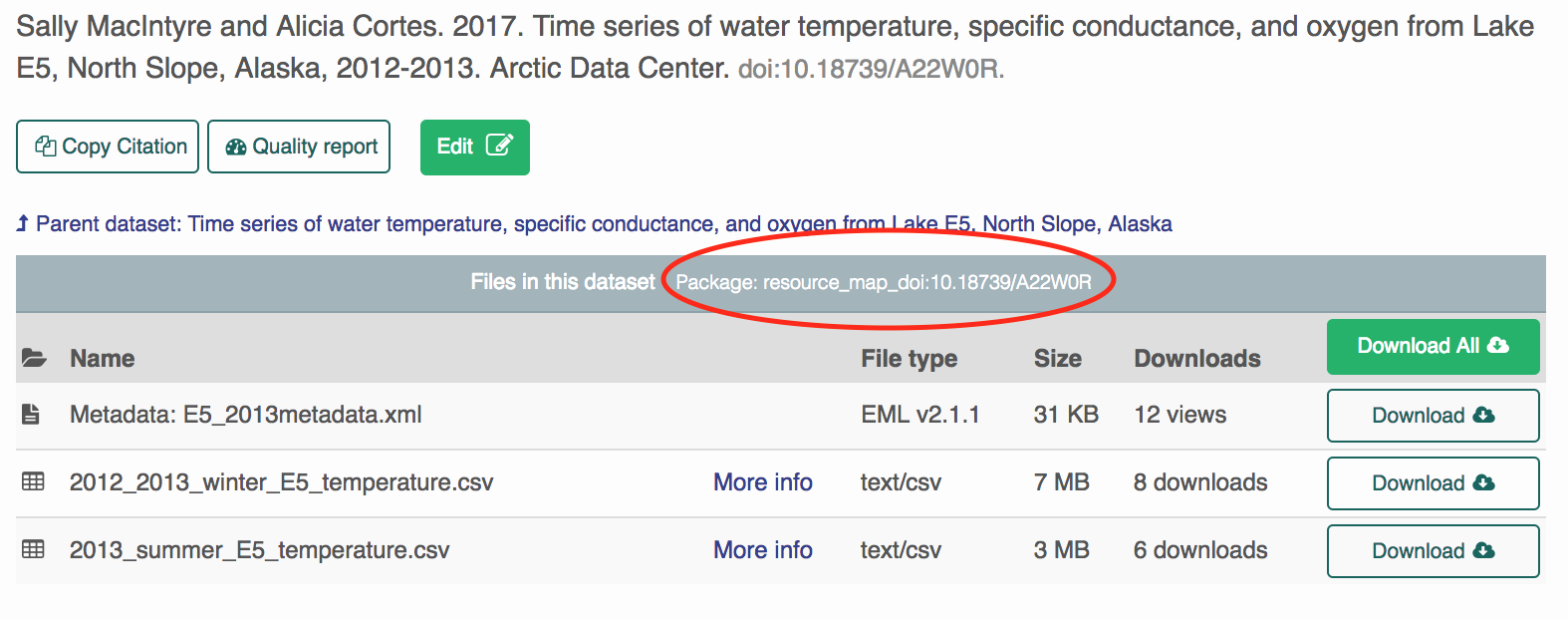
This is a better option (than the first method) if you would like to prefix your data files (in addition to metadata files) with the DOI identifier.
All we need to specify is the package resource map. Copy-paste this into R.
Optional arguments:
-
prefix_file_names- prepends the DOI identifier to each file -
download_column_metadata- downloads column metadata to csv files -
convert_excel_to_csv- attempts to convert Excel workbooks in the package to csv files. Not recommended if the csv files are already present in the package -
download_child_packages- downloads any “Nested” Data Packages associated with the Data Package
download_package(mn,
"resource_map_doi:10.18739/A22W0R",
download_directory = "/home/dmullen/downloads",
prefix_file_names = TRUE,
download_column_metadata = TRUE,
convert_excel_to_csv = FALSE,
download_child_packages = TRUE)Download multiple Data Packages
We can download multiple data packages by specifying multiple resource maps to the function download_packages(). Note the ‘s’!
Tip If you want to download Data Packages that are nested under a common Data Package, you can use the common “parent” resource map rather than copying-pasting multiple resource maps into R.
download_packages(mn,
c("resource_map_doi:10.18739/A21G1P", "resource_map_doi:10.18739/A2RZ6X"),
"/home/dmullen/downloads",
prefix_file_names = TRUE,
download_column_metadata = TRUE)Switching between DataONE/LTER Member Nodes
Suppose we want to download this package from LTER: https://search.dataone.org/#view/https://pasta.lternet.edu/package/metadata/eml/knb-lter-ntl/276/13
We first need to tell R that we are changing Member Nodes. The fastest way of doing this is to reset your mn with the guess_member_node() function.
Note: This function accepts metadata and resource map PIDs or DataONE package URLs as inputs.
# The following three calls return the same member node (mn)
# Metadata pid input:
mn <- guess_member_node("https://pasta.lternet.edu/package/metadata/eml/knb-lter-ntl/276/13")
# Resource map pid input:
mn <- guess_member_node("doi:10.6073/pasta/97c6d83a5f6a0a065ef3209fcb491b6e")
# Package URL input:
mn <- guess_member_node("https://search.dataone.org/#view/https://pasta.lternet.edu/package/metadata/eml/knb-lter-ntl/276/13")
# We can now use any of the above functions:
get_eml_attributes_url(mn, "https://search.dataone.org/#view/https://pasta.lternet.edu/package/metadata/eml/knb-lter-ntl/276/13")